Simple example
This example pipeline showcases the some basic functionality of a (Nipype-based) Porcupine pipeline, which defines a T1-weighted structural scan as input, performs skullstripping on it, and writes the result to disk.
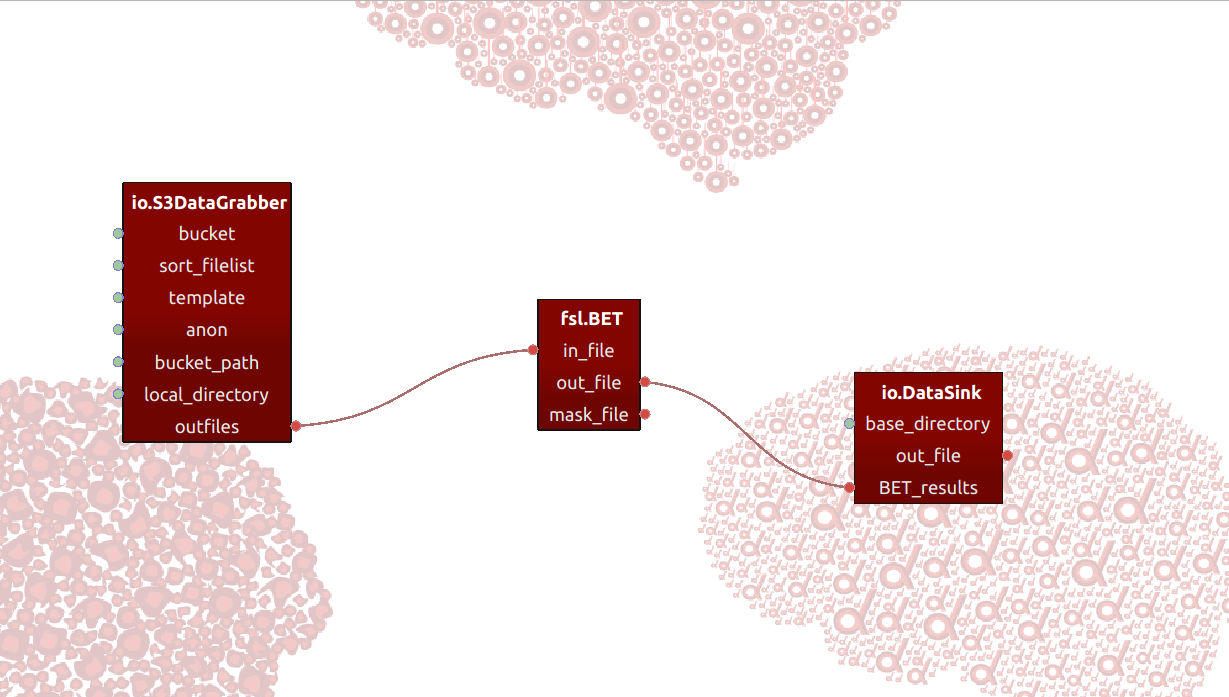
This pipeline, like all the example pipelines we provide, defines the input-data as data from an openneuro Amazon repository (called an “S3 bucket”). In the first node, we define the input as a single T1-weighted structural scan from the ds-000101 dataset from the openneuro repository. Then, we connect this to a skullstripping node (performed by FSL BET), and finally write the skullstripped data to disk in the last node.
The nodes in the pipeline are all based on Nipype-interfaces. Nipype provides
a comprehensive set of interfaces for different neuroimaging software packages,
in addition to more generic functionality relating to file I/O and pipeline-specific
operations. In this example pipeline, we specifically use Nipype’s S3DataGrabber
interface in our first node to download a single T1-weighted file from an
openneuro repository, which we subsequently connect to the Nipype-based FSL
BET node. The result of the skullstrip operation by the BET-node is then written
to disk using the DataSink node from Nipype.
Check out the Porcupine-generated Python-script, Porcupine-pipeline, and associated Dockerfile in our Github repository or download the files directly below.